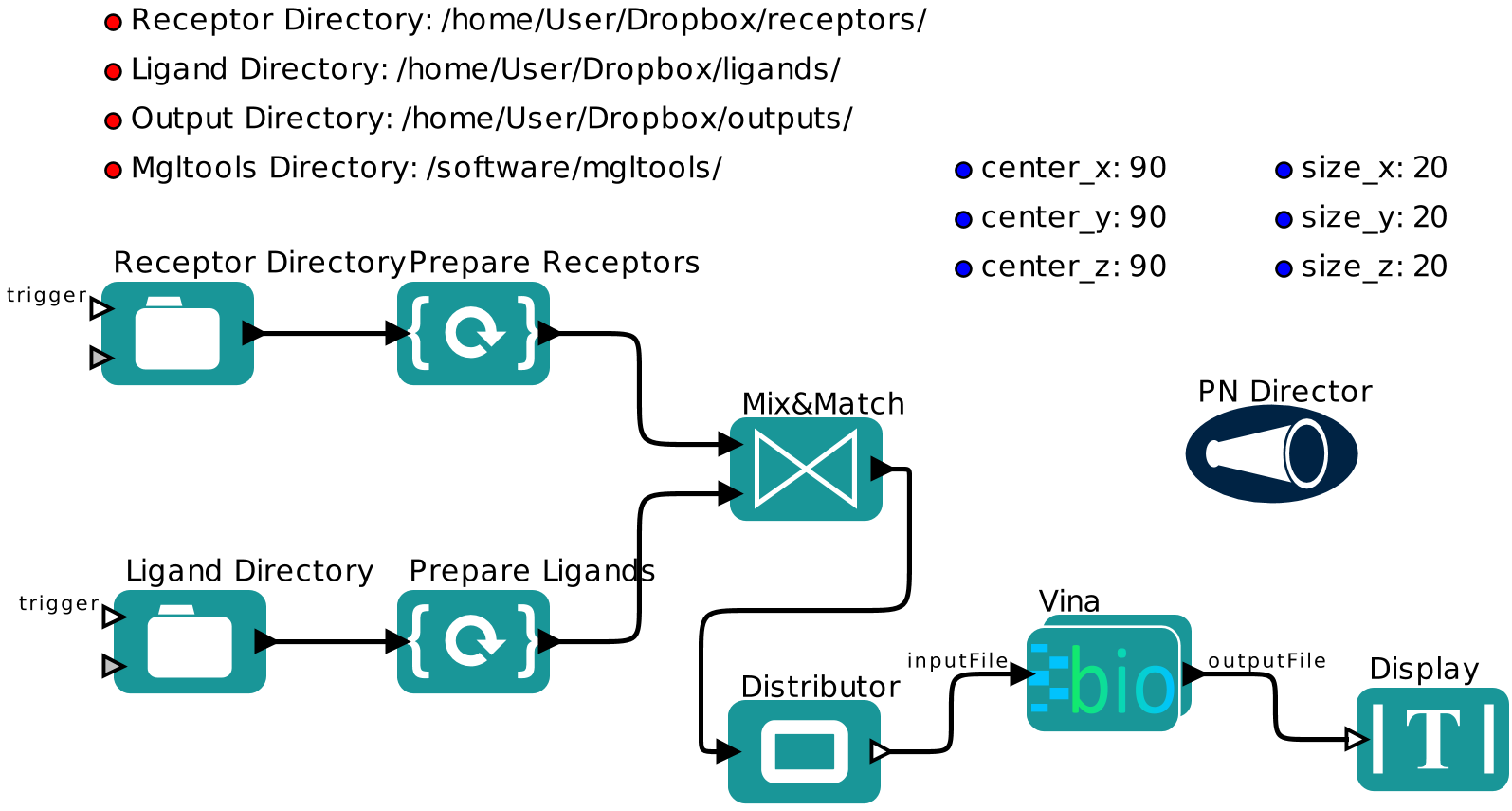
Description
Required Software
biokepler 1.0 or above
MGLTools 1.5.6
Overview
Usage
Parameters
rd Full path to the directory which contains all the receptors of interest.Receptor and ligand directories should be different.
ld Full path to the directory which contains all the ligands of interest.Receptor and ligand directories should be different.
od Full path to the directory where all the docking outputs will be stored.
mgld Directory where the MGLtools are installed. For example, "/home/users/mgltoos/".
center_x The center (x-axis) of the grid box.
center_y The center (y-axis) of the grid box.
center_z The center (z-axis) of the grid box.
size_x The length of the grid box extended from the x-axis center.
size_y The length of the grid box extended from the y-axis center
size_z The length of the grid box extended from the z-axis center.